The CRISPR-Cas system is an acquired immune defense system in prokaryotes against foreign bacteriophage invasion. Based on the composition of Cas effector proteins, the CRISPR-Cas system is classified into Class 1 and Class 2. The effectors of Class 1 systems (further divided into I, III, and IV types) consist of effector complexes with multiple subunits, while Class 2 systems (II, V, and VI types) have single effector proteins. Among these types, the III-type CRISPR-Cas system is of particular interest due to its intricate immune mechanism and cleavage of invader RNA and DNA. The III system recognizes invading RNA molecules and further divides into subtypes III-A to III-F.
Makarova et al. reported the III-E type CRISPR-Cas system in 2019 through bioinformatics analysis, where 4 Cas7 domains and a small subunit similar to Csm2/Cmr5 (Cas11) fuse into a large protein named gRAMP. In the gene cluster of the III-E type CRISPR-Cas system, the characteristic Cas10 of the III system and csm4/cmr5 genes are missing, indicating differences in its immune mechanism. Subsequent studies revealed that the III-E type system can process precursor crRNA into mature crRNA and cleave target RNA at intervals of 6 nucleotides, without RNA off-target effects, making it a potentially highly specific RNA editing tool. Interestingly, the effector protein gRAMP of the III-E type forms a stable complex with the caspase-like protease TPR-CHAT (named Craspase), speculated to be an activated protease recognized by the target RNA through the III-E type effector protein. However, the protease activity remains unconfirmed, and the substrate of TPR-CHAT is also unknown. Furthermore, the immune mechanism of the III-E type CRISPR-Cas system remains a puzzle.
On October 27, 2022, Professor Feng Yue's research group at Beijing University of Chemical Technology collaborated with Professor Yang Maojun's research group at Tsinghua University to publish a research paper titled "Target RNA activates the protease activity of Craspase to confer antiviral defense" online in Molecular Cell. This work reported a series of structures of gRAMP-crRNA from Candidatus "Scalindua brodae" and Craspase, revealing that the cognate target RNA (CTR) induces significant conformational changes in the TPR-CHAT of Craspase. It also unveiled that CTR activates the protease activity of Craspase to cleave the substrate protein Csx30, triggering abortive infection in bacteria as an antiviral strategy of the III-E type system.

The research team first identified that in the gRAMP-crRNA and gRAMP-crRNA-TR (target RNA) structures, D547 in gRAMP and D698/D806 are responsible for cleavage at sites 1 and 2 in the target RNA, respectively. It is noteworthy that the insertion domain in the Cas7.4 domain of gRAMP shows poor density in the electron microscopy data. Mutants lacking the insertion domain exhibit a loss of target RNA cleavage activity and weaker pre-crRNA processing activity. This suggests that the insertion domain may be involved in precursor crRNA processing of gRAMP, which differs from gRAMP from the Desulfonema ishimotonii species, indicating potential mechanistic differences among type III-E effector proteins from different species.
Therefore, the research focus shifted to Craspase to investigate whether the proteolytic activity of TPR-CHAT can be activated and what the proteinase targets are. The team proceeded to analyze the structures of Craspase in four states: alone, and bound to TR/CTR/NTR (non-cognate target RNA). Structural comparisons revealed that binding of CTR (targeting non-self RNA) induces significant conformational changes in regions of TPR-CHAT, including the active site, whereas TR/NTR does not induce such changes. This indicates that the proteolytic activity of TPR-CHAT in Craspase may be activated by binding to CTR.
In order to search for the substrate of TPR-CHAT, the research group analyzed the gene cluster of type III-E. Typically, there are three conserved accessory genes in this cluster: Csx30, Csx31, and a Sigma factor E (σE) encoding SbRpoE. Protease activity experiments showed that in the presence of CTR, Craspase can cleave Csx30 but not SbRpoE. Moreover, the protease activity of Craspase on Csx30 cannot be activated in the presence of TR or NTR. Since Csx31 alone could not be purified to a homogeneous state, no enzymatic cleavage detection was performed on the individual Csx31 protein. Additionally, they also found that Csx30, Csx31, and SbRpoE can form a complex, and the activated Craspase can cleave the Csx30 protein within this complex.
To investigate the biological significance of Csx30 cleavage activated by CTR, the research group transformed genes encoding gRAMP, TPR-CHAT, Csx30, Csx31, and SbRpoE, as well as a synthetic CRISPR array containing sequences for dsDNA bacteriophage lambda (λ) gene transcripts, into Escherichia coli. They conducted experiments on λ phage infection and bacterial growth induction inside the bacterial cells. The experimental results indicated that this system could confer E. coli with resistance to λ phage through a stuttering infection mechanism. Moreover, Craspase recognition of CTR and subsequent cleavage of Csx30 were found to be cytotoxic to the cells, and the cleaved segments of Csx30, when co-expressed with Csx31 and SbRpoE in E. coli, inhibited the growth of E. coli.
Based on the above research, the research group proposed a molecular mechanism model of the III-E immune system, which indicated that TPR-CHAT in Craspase itself is in an inactive state, and the RNA transcribed from phage induces a conformational change in TPR-CHAT by binding to Craspase, activating its activity. TPR-CHAT recognizes the substrate Csx30 and cleaves it, thus triggering bacterial stalling infection in the presence of Csx31 and SbRpoE (Figure 1). However, the detailed mechanism of cell death induced by the cleavage of Csx30 in the presence of Csx31 and SbRpoE remains to be further studied.
Doctoral students Liu Qian and Wang Hao from Beijing University of Chemical Technology, Postdoctoral Zhang Laixing from Tsinghua University, master's student Xiu Yu, Huang Ling, and Gao Zhengyu are the co-first authors of this paper. Professor Feng Yue is the corresponding author and Lead Contact of this paper, Professor Yang Maojun from Tsinghua University and Dr. Zhang Laixing are the co-corresponding authors of this paper. Beijing University of Chemical Technology is the first completing unit.
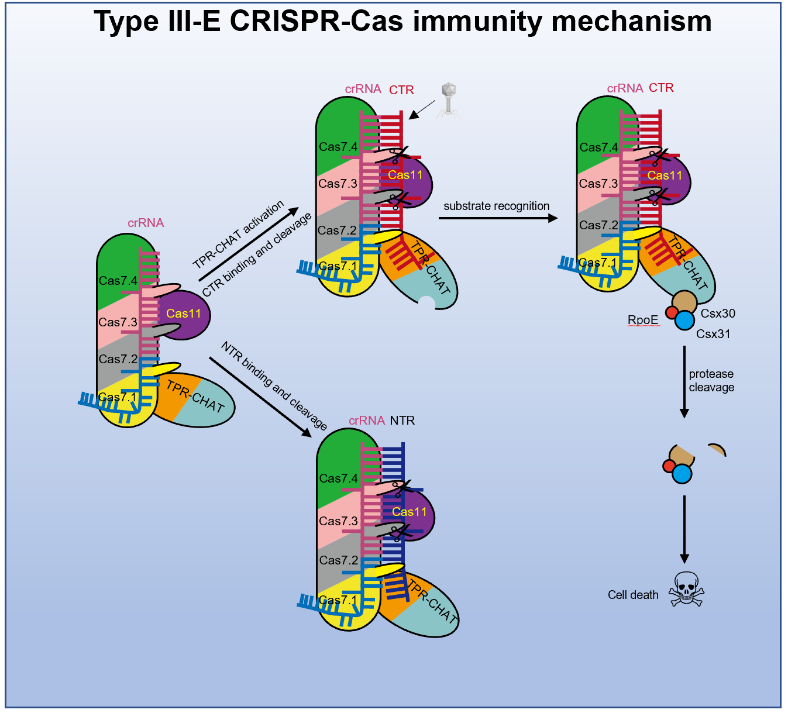
Figure 1. Model of the immune mechanism of the type III-E CRISPR-Cas system.
References:
1.Makarova, K.S. et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol18, 67-83 (2020).
2.Ozcan, A. et al. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature597, 720-725 (2021).
3.van Beljouw, S.P.B. et al. The gRAMP CRISPR-Cas effector is an RNA endonuclease complexed with a caspase-like peptidase. Science373, 1349-1353 (2021).
4.Kato, K. et al. Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex. Cell185, 2324-2337 e2316 (2022).


