In the world of microorganisms, the struggle between bacteria and phages is like an arms race that has been going on for a long time, and they have evolved over a long period of evolutionary history to fight against each other. In order to resist phage infection, bacteria have evolved a variety of anti-phage defense systems, such as the CRISPR-Cas system, etc., and in turn, phages are more powerful than phages, encoding many anti-defense proteins represented by Anti-CRISPR (Acr) proteins.2023 In February 2023, Feng Yue's group collaborated with the University of California, San Francisco's Josef K. K. Kohler and the University of California, San Francisco, to develop a new anti-phage defense system, which is called the “anti-CRISPR (Acr) protein. In February 2023, Feng Yue's group collaborated with Joseph Bondy-Denomy's group at the University of California, San Francisco, to publish an article in Cell reporting the discovery of the phage anti-defense protein Acb2 (Anti-CBASS2) and its mechanism as a sponge adsorption signaling molecule. On October 30, 2024, Yue Feng's group and Joseph Bondy-Denomy's group collaborated again and published a research paper titled Single phage proteins sequester signals from TIR and cGAS-like enzymes in Nature, revealing that Tad1, Tad2, and Acb2 are “super sponges” with two different binding pockets, thus bringing the current discovery of sponge proteins to the forefront of the research. The paper reveals that Tad1, Tad2 and Acb2 are “super sponges” with two different binding pockets, which unifies the only three phage sponges discovered so far as “super sponges” with multiple binding pockets, and establishes a new paradigm in the study of sponge proteins.

Feng Yue's group firstly verified the ability of Tad1 and Tad2 to bind other cyclic oligonucleotides through in vitro biochemical experiments, and found that in addition to the binding effect on 1′-2′-gcADPR/1′′-3′-gcADPR, they can also bind a variety of cyclic dinucleotides or cyclic trinucleotides. In order to reveal the mechanism by which Tad proteins bind signaling molecules, Feng Yue's group resolved the crystal structures of Tad proteins in complex with a variety of cyclic oligonucleotides. They found that the Tad1 protein exists as a hexamer instead of a dimer as previously reported, and that Tad1 acts like a “molecular spaceship” with two “isolation compartments” that can form two separate pockets to isolate the two CTNs, and that Tad1 uses the same binding mechanism as the gcADPR molecule. Tad2 is a tetramer that binds to two CDNs and two gcADPR molecules at the same time (Figure 1). In addition, by quantitatively analyzing the binding affinity of Tad proteins to cyclic oligonucleotides, Feng Yue's group found that Tad1 has a strong binding affinity for common CBASS defense system signaling molecules, such as cA3, 2′,3′-cGAMP, and 3′,3′-cGAMP, whereas Tad2 does not bind to CTNs, but only to CDNs. even more interestingly, they found that A homolog of Tad2 (HgmTad2) has a binding affinity for cGG as high as 24 pM, which is the strongest cGG-binding protein with the highest affinity reported so far, and is also much higher than that of sponge proteins for signaling molecules reported so far. In bacteria, cGG is the most widespread CDN, which acts as a “key hub” in regulating bacterial growth and behavior in various aspects including motility, virulence, biofilm formation, and cell cycle progression. cGG is the first phage antidefense protein targeting cGG, and it is expected to provide ideas for the future study of cGG-related signal transduction pathways. signaling pathways. Subsequently, Bondy-Denomy's group verified that Tad proteins have in vivo activity to inhibit the CBASS system as well as the Thoeris system by anti-phage experiments.
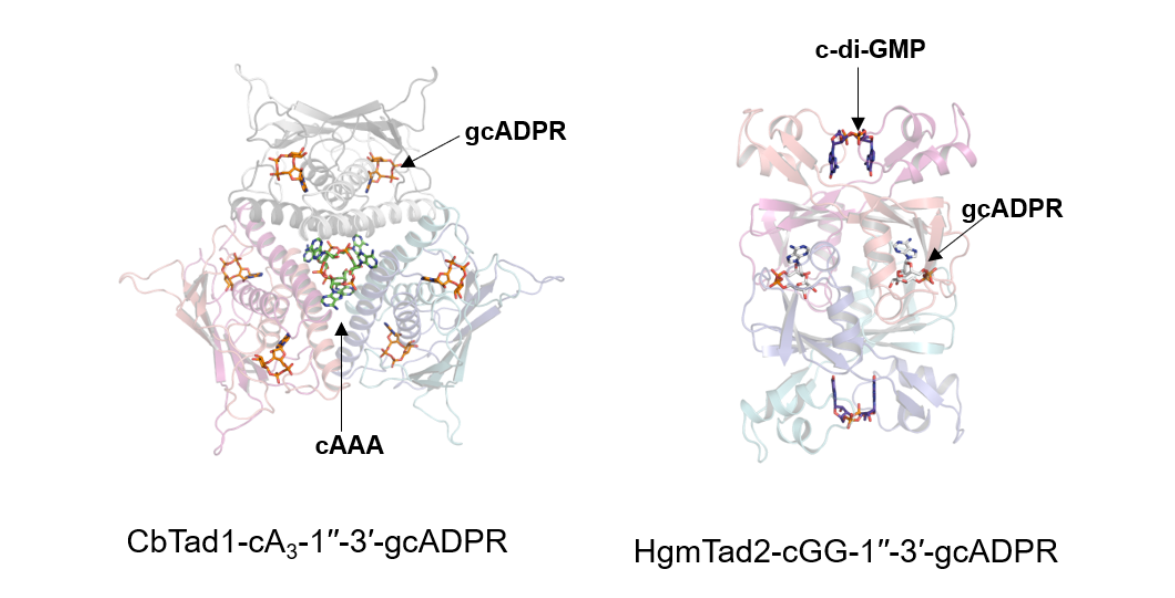
Figure 1 Structures of CbTad1-cA3-1′′-3′′-gcADPR and HgmTad2-cGG-1′′-3′′-gcADPR
In conclusion, this paper and the series of work on Acb2 by Feng Yue's group have confirmed that the only three known sponges, Acb2, Tad1 and Tad2, are “super sponges” with multiple binding sites to isolate signaling molecules, which suggests that anti-defense proteins with multiple binding sites may be widely available. This finding suggests that anti-defense proteins with multiple binding sites may exist widely, thus establishing a new paradigm for the study of phage anti-defense “sponges”.
Dr. Dong Li, a PhD candidate from Beijing University of Chemical Technology, Dr. Yu Xiao from Institute of Botany, Chinese Academy of Sciences, Dr. Iana Fedorova from University of California, San Francisco, Dr. Weijia Xiong, a PhD candidate from Beijing University of Chemical Technology, and Dr. Yu Wang, a Master's candidate, and graduated PhD student, Xi Liu, are the co-first authors of this paper, while Prof. Yue Feng from Beijing University of Chemical Technology and Prof. Joseph Bondy-Denomy from University of California, San Francisco are the co-first authors. Prof. Feng Yue of Beijing University of Chemical Technology and Prof. Joseph Bondy-Denomy of University of California, San Francisco are the co-first authors of this paper. Beijing University of Chemical Technology is the first organization.
Original link: https://www.nature.com/articles/s41586-024-08122-4
Yue Feng is a professor and doctoral supervisor at Beijing University of Chemical Technology, and a recipient of the National Excellent Youth Fund (2018). He mainly conducts research on the mechanism of microbial-host immune system interactions by means of biochemistry and molecular biology, structural biology and cell biology. He has published a total of 53 SCI papers, including 30 papers with corresponding authors (including co-authors) in Nature (2), Cell, Mol Cell (3), Nat Chem Biol (3), PNAS, Nat Plants, Nat Commun (2), Nucleic Acids Res, and other internationally renowned journals. He has been in charge of a number of national and provincial projects. He has been awarded the honors of Young Beijing Scholar (2024), National Young Post Master (2020), Beijing Outstanding Young Talent (2020), Second Prize of Beijing Science and Technology Progress Award (2018), China's Top 10 Emerging Science and Technology Figures (2018), and Beijing Science and Technology Rising Star (2019).
