On March 20, 2025, Feng Yue's group at the State Key Laboratory of Green Biomanufacturing and Beijing University of Chemical Technology, in collaboration with Joseph Bondy-Denomy's group at the University of California, published a research paper in Cell titled Jumbo phage killer immune system targets early infection of nucleus-forming phages. The study identifies the first defense system against giant phages in bacteria, the “jumbo phage killer” (Juk for short), and reveals its mechanism of operation, providing direction for the study of bacterial anti-giant phage systems.
As a virus that infects bacteria, phages have long evolved a series of delicate mechanisms to ensure that they can efficiently utilize host cell resources for their own survival. In order to resist phage infection, bacteria have developed complex and diverse anti-phage defense mechanisms. At the same time, the “arms race” between phages and bacteria has driven phages to develop a variety of counter-defense mechanisms, making the interactions in this microbial world increasingly complex. With the in-depth study of these defense and counter-defense mechanisms, scientists have gradually revealed how bacteria respond to phage challenges and phage countermeasures through a variety of defense systems, demonstrating the amazing diversity and adaptability of this microscopic world. However, although hundreds of phage-resistant defense systems have been reported, bacterial defense mechanisms against phiKZ-like jumbo phages (i.e., phages with genomes >200 kb) remain understudied.
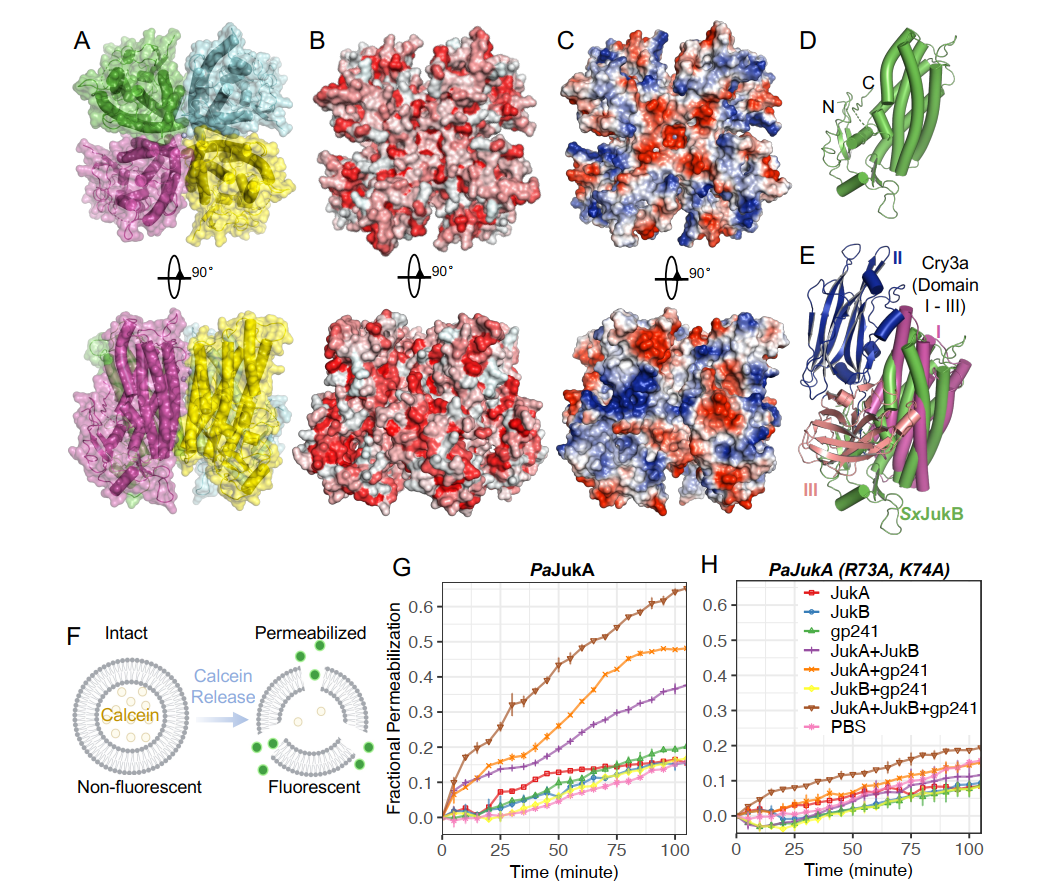
In this study, the research team first identified that the Juk system consists of two parts, including JukA and JukB. they found that JukA is a sensor protein that rapidly recognizes the phage protein gp241, which is expressed early in the phage infection process. at the early stage of infection, JukA rapidly binds to gp241 and localizes to the site of phage infection, which then activates the JukB effector role.3 What is the function of JukB? Feng Yue's team successfully resolved the structure of the JukB protein by X-ray crystallography: the structure is a tetramer and presents a negatively charged pore-like structure in the center. Interestingly, the structure of JukB is highly similar to that of the Bacillus thuringiensis pore-forming toxin Cry3A, which functions by forming holes in the cell membranes of insect intestinal cells in order to disrupt cellular functions, thus leading to insect death. It is worth mentioning that, in order to be protected from bacterial CRISPR-Cas and other nucleases, the phiKZ family of giant phages assembles a lipid-coated early phage infection (EPI) vesicle and a protein-like nucleoid structure to protect its genome during bacterial infection. Based on the structural similarity between JukB and pore-forming toxin, Feng's team wondered whether JukB would similarly destabilize EPI vesicles in the same way that Cry3A destabilizes the membranes of insect intestinal cells. Subsequently, Feng's team further determined that JukA, JukB and phage protein gp241 could form a ternary complex through in vitro biochemical experiments, and verified that the complex could effectively destroy model liposomes in vitro through liposome-calcium xanthophyll leakage experiments, which suggests that JukB may similarly destabilize EPI vesicles. By disrupting EPI vesicles, JukB can expose phage DNA to nuclease in the bacterial cytoplasm, thereby inhibiting phage gene replication and expression. Finally, the collaborative team demonstrated that the Juk system can effectively inhibit most phage gene expression, phage DNA replication and phage nuclear assembly.
Figure Tetrameric structure of JukB and its disruption of early phage infection (EPI) vesicles
In summary, this paper identifies and resolves the mechanism of action of a novel bacterial anti-phage system, Juk. juk, a widespread immune system, specifically detects phiKZ-like giant phages early in infection, blocks phage replication, and allows host cells to continue dividing, which contrasts with the mostly abortive infection properties of the immune system. The “arms race” between phages and bacteria has been a hot topic in microbiology. The discovery and study of the Juk immune system demonstrates the unique defense capabilities of bacteria against complex viruses such as giant phages, which not only helps us to better understand how bacteria respond to phage attacks, but also may provide important clues for the development of novel antimicrobial strategies.
Dr. Yuping Li from UCSF and Linlin Guan, a master's student from Beijing University of Chemical Technology, are the first and second authors of this paper, respectively, and Professor Yue Feng from Beijing University of Chemical Technology, Professor Joseph Bondy-Denomy and Dr. Yuping Li from UCSF are the co-corresponding authors of this paper.
Author introduction:
Yue Feng is a professor at Beijing University of Chemical Technology (BUCT) and a supervisor of Ph.D. He graduated from Tsinghua University with a Ph.D. in 2013 and joined BUCT in the same year. Dr. Yue Feng's group has been conducting research on the structure and function of proteins related to the interaction between microbes and host immune system by means of biochemistry and molecular biology, structural biology and cell biology for many years. He has published a total of 32 SCI papers with corresponding authors (including co-authors) in Nature (2), Cell (2), Nat Chem Biol (3), Mol Cell (3), PNAS, Nat Plants, Nat Commun (2), Nucleic Acids Res and other journals. He has been awarded the National Young Post Master (2020), Young Beijing Scholar (2024), the 19th Youth Science Award for Colleges and Universities of Higher Education of the Fok Ying Tung Education Foundation (2024), Beijing Outstanding Young Talent (2020), the Second Prize of Chinese Medical Science and Technology Award (2024), China Top 10 Emerging Science and Technology Figures (2018), Beijing Science and Technology Rising Star (2019), and the Beijing Higher Education Young Teacher Outstanding Instructor Award of Basic Teaching Skills Competition (2023), etc.