Recently, the Wu Bian/Cui Yinglu team published a research paper titled “Glycolysis-compatible urethanases for polyurethane recycling” in the journal Science. Utilizing an AI-driven microbial enzyme resource mining strategy, the research team successfully discovered a novel urethane-degrading enzyme, AbPURase. This enzyme exhibits outstanding tolerance to organic solvents, maintaining exceptionally high catalytic activity and stability even in high-concentration alcoholysis reaction media. It demonstrates high compatibility with industrial polyurethane alcoholysis processes. This achievement marks the first successful large-scale biological depolymerization of polyurethane under industrial conditions, providing an efficient and sustainable new pathway for the green recycling of polyurethane plastics.
As the world's sixth most widely used polymer, polyurethane (PU) plastics exceed 30 million tons in annual production. Leveraging their outstanding physicochemical properties, they are extensively applied across multiple sectors of the national economy. Applications span building insulation and thermal materials, consumer goods (e.g., mattresses, sofa padding, refrigerator and air conditioner insulation layers), and transportation vehicles (e.g., automotive and aircraft seat components). However, as its application scope and production scale continue to expand, the environmental pressure from polyurethane waste is intensifying. Developing resource recovery technologies for waste polyurethane plastics has become an urgent priority. As a polymeric material, polyurethane is primarily produced through the reaction of isocyanate with polyether polyols. Theoretically, depolymerization through chemical methods like hydrolysis could recover the original monomers for recycling. However, polyurethane's highly cross-linked three-dimensional network structure makes depolymerization conditions demanding and hinders efficient, complete monomer recovery, severely limiting the industrialization of closed-loop recycling.
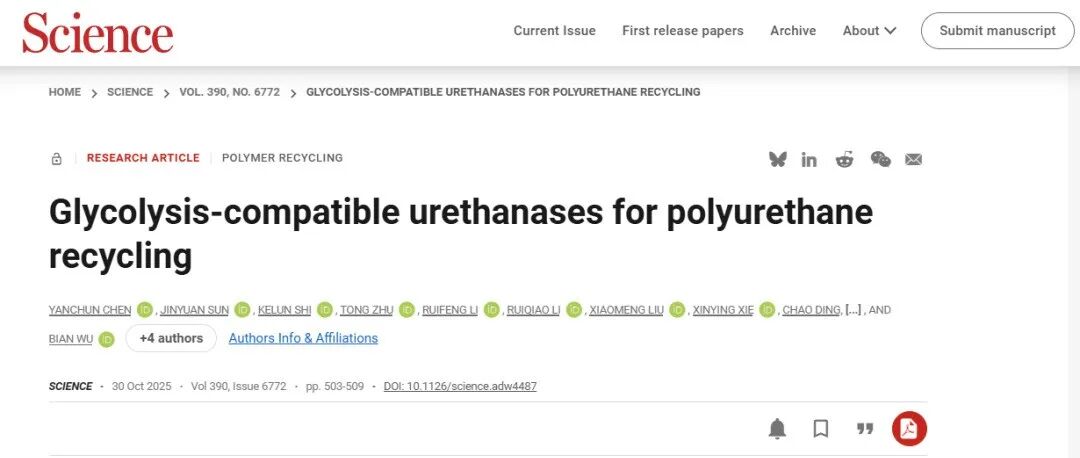
Achieving enzymatic hydrolysis of polyurethane hinges on developing ureases capable of efficiently hydrolyzing urethane bonds. Yet, existing EC enzyme classification systems lack clear categorization for such ureases. The few known ureases capable of hydrolyzing aromatic urethane bonds are traditionally classified as lipases or amidases in bioinformatics systems. Therefore, the research team leveraged their independently developed protein pre-training model Pythia (The Innovation 2025, 100750, https://pythia.wulab.xyz), combined supervised and self-supervised learning to develop GRASE (GNN-based Recommendation of Active and Stable Enzymes), a graph neural network-based enzyme discovery tool. This tool predicts protein stability by estimating the likelihood between protein sequences and structures. It utilizes graph neural networks to extract local microenvironment and overall structural information of amino acids within catalytic pockets. By calculating the cosine similarity between feature vectors of different catalytic pockets in high-dimensional space, it measures their functional similarity, enabling precise inference of enzyme substrate specificity and reaction preferences.
GRASE Model Architecture Diagram
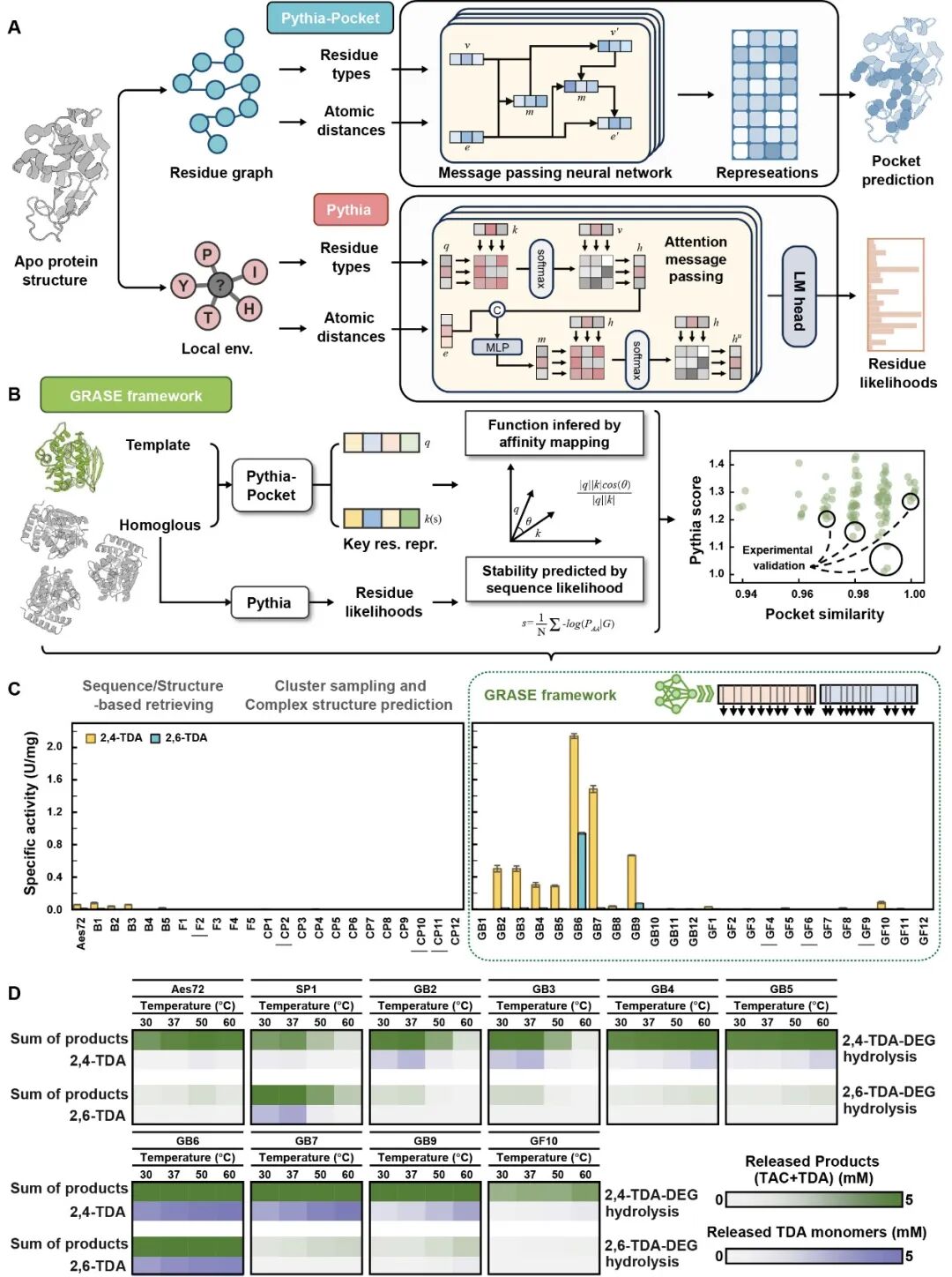
Using the GRASE tool, the team successfully identified multiple enzymes with carbamate degradation activity from metagenomic data. Among these, the AbPURase enzyme from Alicyclobacillus sp. demonstrated outstanding properties across all aspects. This enzyme features a V-shaped active pocket stabilized by a compact hydrophobic core and multiple proline residues within a lid domain. This unique architecture not only facilitates substrate binding but also effectively blocks solvent penetration into the active site, ensuring AbPURase maintains structural and functional integrity in high-concentration organic solvents.
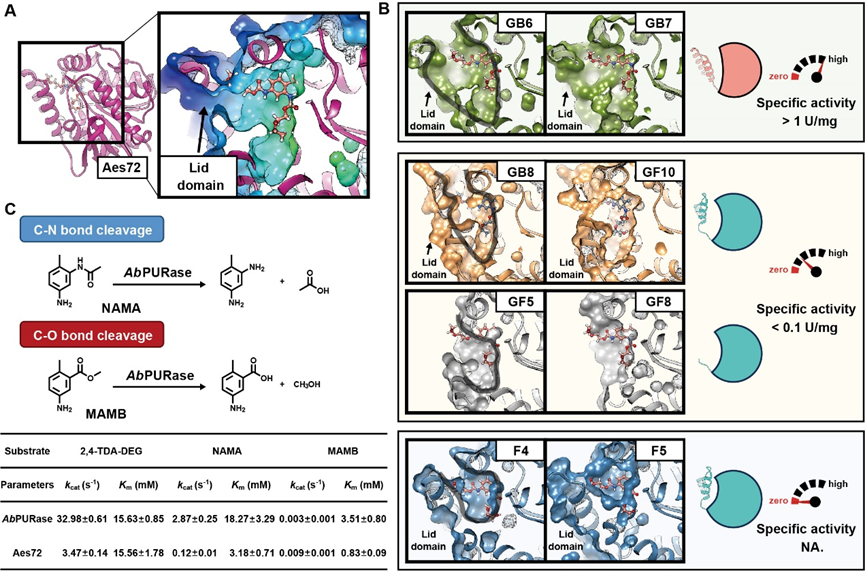
Structural Analysis of a Newly Developed Urease
Benefiting from its unique natural structural properties, the AbPURase enzyme exhibits catalytic activity up to 465 times higher than previously reported ureases in high-concentration diethylene glycol environments during industrial alcoholysis processes, demonstrating exceptional catalytic efficiency and strong organic solvent tolerance. In kilogram-scale commercial polyether polyurethane foam degradation experiments simulating industrial reaction conditions (substrate loading 500 g/L), AbPURase achieved a 98.6% substrate conversion rate within 8 hours, with recovery rates for the degraded monomer TDA and the alcoholysis agent DEG reaching 94.7% and 98.5%, respectively. Furthermore, AbPURase demonstrated outstanding operational stability, maintaining high degradation activity through multiple cycles, indicating its potential for industrial applications.
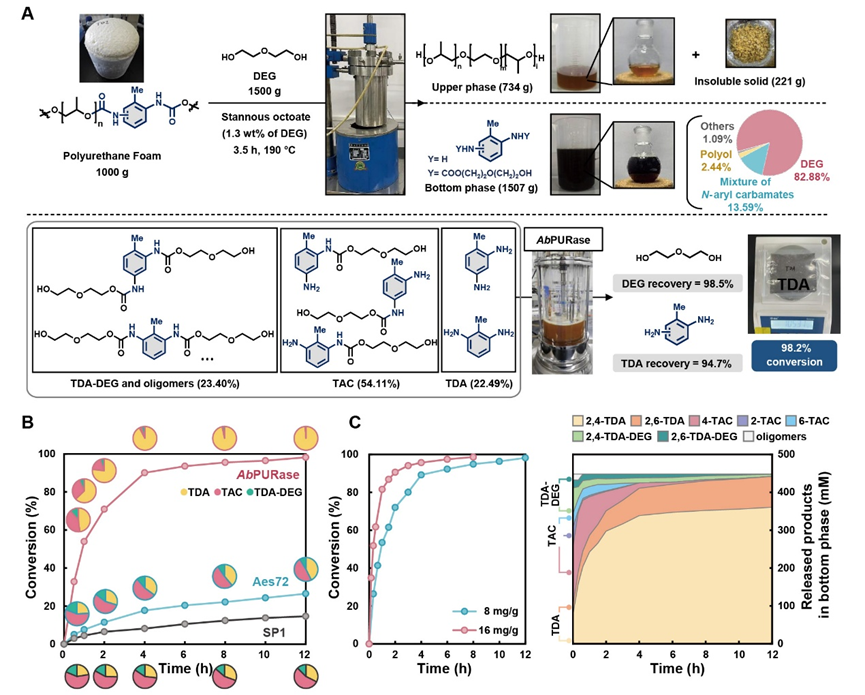
Enzymatic Alcoholysis of Commercial Polyurethane
Overall, the hundreds of millions of known protein sequences harbor rich biological patterns and evolutionary information. However, traditional enzyme discovery methods typically rely on sequence similarity or coarse-grained enzyme classification codes for screening, leading to the systematic omission of numerous enzyme resources with low sequence similarity but similar functions. The GRASE system developed by the research team effectively overcomes the limitations of traditional sequence analysis methods. By leveraging deep learning to capture high-dimensional features of local protein structures, it efficiently uncovers the “dark matter” of microbial enzyme resources hidden in nature. This research not only provides a solution with industrial application potential for plastic recycling but also advances the intelligent development of artificial intelligence in the field of industrial enzyme discovery.
This project was jointly completed by Beijing University of Chemical Technology and the Institute of Microbiology, Chinese Academy of Sciences. Professor Wu Bian and Researcher Cui Yinglu served as co-corresponding authors, with Special Research Assistant Chen Yanchun and doctoral candidates Sun Yuen and Shi Kelun as co-first authors. The research received funding from the National Key R&D Program, the National Science Fund for Distinguished Young Scholars, and the National Science Fund for Excellent Young Scholars.
Profile of Professor Wu Bian
Wu Bian, Professor at the School of Life Sciences and Technology, Beijing University of Chemical Technology, is a recipient of the National Science Fund for Distinguished Young Scholars (2023). He serves as the principal investigator for a National Key R&D Program project and as a member of its guideline expert panel. Additionally, he chairs the Enzyme Engineering Committee of the Chinese Society for Microbiology. His research focuses on biocatalysis, particularly on enzyme component discovery, mechanism elucidation, and engineering modification. By integrating artificial intelligence into enzyme engineering, he has advanced the design of biomacromolecules. His research has been published in prestigious journals including Science, Nature Catalysis, and Nature Chemical Biology. Multiple technologies developed by his team—such as enzymatic assembly of peptide drugs, biosynthesis of β-amino acids, and sequential fermentation of azetidine drug intermediates—have achieved successful industrial applications. He has received the First Prize of Hebei Provincial Science and Technology Progress Award, the Zhonghe Young Peptide Scientist Award, and the Outstanding Young Enzyme Engineer Award from the Chinese Society for Microbiology.
